Abstract
The organization of the human genome in space and time is critical for transcriptional regulation and cell fate determination. However, robust methods for tracking genome organization or genomic interactions over time in living cells are lacking. Here, we developed a multicolor DNA labeling system, ParSite, to simultaneously track triple genomic loci in the U2OS cells. The tricolor ParSite system is derived from the T. thermophilus ParB/ParSc (TtParB/ParSc) system by rational design. We mutated the interface between TtParB and ParSc and generated a new pair of TtParBm and ParSm for genomic DNA labeling. The insertions of 16 base-pair palindromic ParSc and ParSm into genomic loci allow dual-color DNA imaging in living cells. A pair of genomic loci labeled by ParSite could be colocalized with p53-binding protein 1 (53BP1) in response to CRISPR/Cas9-mediated double-strand breaks (DSBs). The ParSite permits tracking promoter and terminator dynamics of the APP gene, which spans 290 kilobases in length. Intriguingly, the hybrid ParS (ParSh) of half-ParSc and half-ParSm enables for the visualization of a third locus independent of ParSc or ParSm. We simultaneously labeled 3 loci with a genomic distance of 36, 89, and 352 kilobases downstream the C3 repeat locus, respectively. In sum, the ParSite is a robust DNA labeling system for tracking multiple genomic loci in space and time in living cells.
Citation: He X, Sun Y, Ma H (2025) ParSite is a multicolor DNA labeling system that allows for simultaneous imaging of triple genomic loci in living cells. PLoS Biol 23(1):
e3003009.
https://doi.org/10.1371/journal.pbio.3003009
Academic Editor: Tom Misteli, National Cancer Institute, UNITED STATES OF AMERICA
Received: June 4, 2024; Accepted: January 9, 2025; Published: January 24, 2025
Copyright: © 2025 He et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All relevant data are within the paper and its Supporting Information files.
Funding: This work was supported by the National Natural Science Foundation of China (No. 2023YFA0913402 to HM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Abbreviations:
DSB,
double-strand break; FROS,
fluorescence repressor operator system; LLPS,
liquid–liquid phase separation; SNR,
signal-to-noise ratio; TAD,
topologically associating domain
Introduction
Chromatin structure and dynamics are important for gene regulation and DNA repair [1,2]. For chromatin structure, many methods have been developed to resolve its fine folding in vitro [3] or in vivo [4]. Chromosome conformation capture [5,6] (3C) and its derived Hi-C [7] have been applied for studies of 3D genome organization extensively. Hi-C data can define genomic interactions or loops, topologically associating domains (TADs), and A/B compartments from a population of cells [7–9]. It is challenging to identify genomic interactions or loops by Hi-C in single cells. DNA FISH such as OligoPaint [10], MERFISH [11,12], Hi-M [13], or ORCA [14] allows for direct visualization of chromatin structures from kilobase to megabase scales in cells. These methods were used to identify genome folding in 3D in fixed cells. Nevertheless, the genome structure is highly dynamic over time in living cells. It is critical to have approaches for visualization or tracking genomic interactions and folding in real-time (the fourth dimension).
Live cell imaging tools aid us to study chromatin dynamics. The fluorescence repressor operator system (FROS) [15,16], CRISPR-based imaging system [17–21], or the ParB/ParS [22–24] system allows us to track genomic loci in living cells. The FROS achieves live cell DNA imaging by the integration of tandem lac operator (LacO) sequences and recruiting its binding partner lac repressor fused with fluorescent protein [15]. At least 20 copies of LacO (typically 100 to 300 copies and up to 100,000 copies) are required for successful DNA imaging but with a low signal-to-noise ratio (SNR) [25]. CuO array or TetO [26] array could be also combined with LacO for multicolor DNA imaging but with a requirement for a minimal of 144 [26] or 28 [27] copies, respectively. The DNA repeat fragment clone and integration for large copies of the FROS array are difficult to tackle. Besides, the integration of multiple copies of FROS sequence may also perturb genome structure and function. For minimal copies of FROS array, low SNR constrains its application in live-cell DNA imaging, in certain cell types with cutting-edge microscopes. CRISPR-based DNA imaging was achieved by targeting the tandem repeats in the human genome using nuclease dead Cas9 (dCas9) fused with the fluorescent proteins along with the cognate sgRNA. CRISPRainbow [20] could be used for simultaneous visualization of 6 genomic loci with high copies of tandem repeats, while CRISPR-Sirius [21] permits labeling the tandem repeats as low as 20 copies. Recently, the non-repetitive DNA was visualized by liquid–liquid phase separation (LLPS)-mediated signal amplification on the genomic locus targeted by a single sgRNA [28]. For CRISPR-based DNA imaging systems, the transient transfection of sgRNA and Cas protein causes heterogenous SNR among cells compared to isolated clones from FROS or other stable cell line-based methods. The heterogenous SNR is a long-standing issue for the multicolor DNA imaging field. The CRISPR-based system has achieved repetitive and non-repetitive DNA imaging with success on a case-by-case basis [17–21,28]. The dCas9 binding to genome DNA may also hinder transcription and stall replication initiation or elongation. Although there are some possible methods to achieve dual- or multicolor DNA labeling through a combination of the above tools, however, a simple and versatile multicolor DNA imaging method from one system is still needed. The ParB/ParS-based ANCHOR systems achieve DNA visualization by integrating sequence from bacteria containing ParS into a genomic locus, which allows the fluorescent protein-fused ParB proteins to spread kilobases around ParS [22]. Recent study also shows that ParS and ParB associate to form nanometer-sized spherical condensates in bacteria [29]. We have previously developed mParSpot based on the ParB-ParS and Noc-NBS systems (another component from bacteria) for tracking pairwise genomic loci in living cells [30]; nevertheless, the mParSpot is primarily a dual-color DNA imaging system.
Here, we developed a multicolor ParSite system based on T. thermophilus ParB/ParS (TtParB/ParSc) system. By mutation of the interface amino acids between TtParB and ParSc, we generated a new pair of TtParBm and ParSm for dual-color DNA imaging. This paper also generated the hybrid of half-ParSc and half-ParSm (ParShybrid, ParSh) which allows for tri-color genomic DNA imaging. Thus, we believe the multicolor ParSite is a robust and sensitive DNA labeling system for exploring genome structure and dynamics in living cells.
Results
Rational design of TtParBm for dual-color DNA labeling
To expand the ParB/ParS-based DNA imaging toolbox in living cells, we rationally designed mutations for ParB protein from Thermus thermophilus (Fig 1A). As we published previously [30], TtParB showed the best SNR for genomic loci-specific DNA imaging among orthogonal ParBs and paralogous Nocs. Interestingly DNA-binding specificity of ParBs can be switched from ParS to NBS by mutations of 4 conserved amino acids [31]. Through protein sequence alignment, we mutated these 4 respective positions in TtParB resulting in TtParBm (TtParB mutant) and also mutated 4 nucleotides in ParSc (ParS consensus) toward to NBS-like sequences, named ParSm (ParS mutant) (Fig 1A).
Fig 1. Generation of TtParB-ParSc mutants for live cell DNA imaging.
(A) AlphaFold models of DBDs from dimeric TtParB (top, purple) and TtParBm (top, red) binding ParSc and ParSm, respectively. The organizations of TtParB or TtParBm and DNA (yellow line) are based on recent structural studies (PDB entry 6T1F and 7OL9). The nucleotide sequences of the ParSc and ParSm are shown below the structure. The red letters indicate the nucleotide differences between ParSc and ParSm. Sequence alignment of α6 and α8 in the DBD of TtParB and TtParBm are shown below. Red dots mean the mutation sites and their locations in the 3D structure are plotted. (B) The schematic of TtParB-8×ParSc or TtParBm-8×ParSm for DNA imaging; 8×ParSc or 8×ParSm is integrated in S1 locus which is 36 kb downstream of C3 repeat (as an endogenous reference locus). (C) Representative images of U2OS-8×ParSc or U2OS-8×ParSm cell line labeled by TtParB-HaloTag or TtParBm-HaloTag, respectively. C3 repeat was labeled by dCas9-GFP/sgRNA-C3. Scale bars, 5 μm for images with the whole cells and 1 μm for zoomed images. (D) Percentage of 8×ParSc (blue) or 8×ParSm (brown) cells labeled by TtParB-HaloTag or TtParBm-HaloTag, respectively, n = 22, 22, 30, 36 cells for each group from left to right. (E) The signal (S)-to-noise (N) ratio of DNA imaging by TtParB-8×ParSc or TtParBm-8×ParSm. The red line indicates the mean value for each group. n = 31 for TtParB-8×ParSc group, 42 for TtParBm-8×ParSm group. The underlying data associated with this figure are available in S1 Data.
To test the labeling efficiency and specificity of TtParB/ParSc and TtParBm/ParSm, we integrate 8 copies of ParSc or ParSm into the S1 locus, 36 kb downstream the C3 locus, a repetitive region (approximately 600 copies) in chromosome 3 (Figs 1B and S1). After transient transfection of TtParB or TtParBm into U2OS-8×ParSc or U2OS-8×ParSm cells, respectively, TtParB could specifically label S1 locus integrated with 8×ParSc and TtParBm with 8×ParSm without cross-reaction (Fig 1C). As statistical data shown in Fig 1D, the labeling efficiency is 100% for TtParB/8×ParSc and 97.22% for TtParBm/8×ParSm. The mean value of SNR is 11.31 for TtParB/8×ParSc and 3.75 for TtParBm/8×ParSm (Fig 1E). To clarify the factors influencing SNR, we quantified the expression level of ParB and ParBm in each cell. The background fluorescence intensity of ParB or ParBm is negatively correlated to the SNR (S2 Fig). Thus, TtParB/8×ParSc and TtParBm/8×ParSm could be utilized for dual-color DNA imaging and we termed this system as dParSite (dual-color ParSite).
Simultaneously labeling a pair of genomic loci by dParSite
To confirm that the dParSite could simultaneously label 2 target sites on a single chromosome, we knocked in 8×ParSm at the S1 locus, 36 kb downstream of the C3 locus (approximately 600 copies) and 8×ParSc at the S2 locus, 89 kb downstream of the C3 locus into U2OS cells. We transfected TtParBm-HaloTag to visualize the S1 locus and TtParB-SNAP to visualize the S2 locus, along with dCas9-GFP/sgRNA-C3 for labeling C3 locus (Fig 2A). As shown in Fig 2B, the S1 spot (8×ParSm in red) and S2 spot (8×ParSc in blue) adjacent to the C3 spot (C3 repeat in green) were visualized simultaneously in the U2OS-8×ParSm-8×ParSc cells. The genomic distance between C3 and S1, S1 and S2, or C3 and S2 is 36 kb, 53 kb or 89 kb, respectively. As shown in Fig 2C, the 2D spatial distances of these loci were measured to range from 80.4 to 829.4 nm with a mean of 332.98 nm for C3 and S1, 48.84 to 826.9 nm with a mean of 308.36 nm for S1 and S2, 145.12 to 1159.88 nm with a mean of 502.39 nm for C3 and S2 from images with 3D projection. We also measured the 3D distance of pairwise genomic loci. It also shows a heterogenous distribution with almost the same pattern as the one from 2D distance (S3A and S3B Fig). To verify the correct localization of the ParSite Tag, we used CRISPR-based labeling of C3 as a reference, which is 36 kb upstream of the S1 site. It will be intriguing to further explore whether CRISPR-based labeling or ParSite labeling affects chromatin topology or their spatial distance. The foci areas from 3D projected images also show heterogeneous distribution (S3C Fig). The labeling efficiency of dParSite is 90.5% (S4 Fig).
Fig 2. Imaging of pairwise genomic loci by the dual-color ParSite system.
(A) The diagram shows the dual-color ParSite system for imaging of pairwise DNA loci. U2OS cells were integrated with 8×ParSm at S1 locus and 8×ParSc at S2 locus resulting in U2OS-8×ParSm-8×ParSc cell lines. TtParBm-HaloTag and TtParB-SNAP along with SgRNA-C3 and dCas9-GFP were co-transfected into cells to image the C3, S1, and S2 loci simultaneously. Combining ParB/ParSc and ParBm/ParSm constitutes the dual color ParSite system. (B) Representative images of S1 and S2 locus in A labeled by ParSite system. Scale bars, 5 μm for images with the whole cells and 1 μm for zoomed images. (C) Box-and-Whisker plots of 2D spatial distances among C3-S1, S1-S2, and C3-S2. n = 38 for each group. Box spans from second to third quartiles. Whiskers represent min to max value, and middle lines represent the position of the median value of the data distribution. The underlying data associated with this figure are available in S2 Data.
GFP-53BP1 foci concurrently associate with 2 adjacent sites labeled by dParSite
53BP1, a key regulator of the DNA repair pathway, forms foci around the double-strand break (DSB) site right after the DNA damage occurs [32]. Here, we introduced DSBs by CRISPR Cas9 nuclease 5 kb downstream of C3 locus and/or 10 kb upstream of S2 locus (Fig 3A), and then examined the colocalization between C3 spot or S2 spot and 53BP1 foci. We have previously shown that C3 locus can be visualized by Cas9 nuclease and truncated guide RNA (11 nucleotides in length), which are sufficient to recognize target sites but lacking cleavage activity [33]. As shown in Fig 3B–3D, 53BP1 foci were associated with C3 when the cleavage site adjacent to C3 repeat; 53BP1 foci were associated with S2 when the cleavage site adjacent to S2; 53BP1 foci were associated with both C3 and S2 when the cleavages occur at both sites. The percentage of cells having C3 locus colocalized with 53BP1 foci is 18%. The percentage is 15.9% for the S2 locus colocalized with 53BP1 foci. The percentage of cells with both C3 and S2 loci colocalized with 53BP1 foci is 66.03% (Fig 3E). When no CRISPR-mediated DNA cleavage is induced, there are no 53BP1 foci colocalized with C3, S1, or S2 loci (S5 Fig). These data suggest 53BP1 forms foci around the DSB sites, and two 53BP1 foci may fuse together when two DSBs occur.
Fig 3. Visualizing the genomic loci with DNA DSBs by the ParSite system in live cells.
(A) The schematic of labeling genomic loci with DNA DSBs using the ParSite system. The 2 red scissors represent 2 cleavage sites mediated by the CRISPR-Cas9 system. (B) Colocalization of the CRISPR-mediated cleavage site adjacent to C3 locus and 53BP1. The cleavage site (scissor) adjacent to the C3 repeat was shown in the diagram. GFP-53BP1 was chosen to indicate the DSB induced by the CRISPR-Cas9/sgRNA system. Scale bars, 5 μm for images with the whole cell and 1 μm for zoomed images. (C) Colocalization of the CRISPR-mediated cleavage site adjacent to the S2 locus and 53BP1. The cleavage site (scissor) adjacent to the S2 locus was shown in the diagram. Scale bars, 5 μm for images with the whole cell and 1 μm for zoomed images. (D) Colocalization of the CRISPR-mediated cleavage at 2 DNA sites and 53BP1. The cleavage sites (scissors) with one close to the C3 repeat and the other close to the S2 locus. Scale bars, 5 μm for images with the whole cell and 1 μm for zoomed images. (E) The percentage of cells grouping in B–D. m1 means only C3 locus colocalized with 53BP1 protein and m2 means only S2 locus colocalized with 53BP1 protein. m1 + m2 means both C3 locus and S2 locus colocalized with 53BP1 protein. n = 17 cells. The underlying data associated with this figure are available in S3 Data.
Imaging promoter and terminator of the APP gene by the dParSite
It has been proposed that the promoter and terminator may interact during active transcription [34]. To examine promoter and terminator interactions, we chose the human Amyloid Beta Precursor Protein (APP) gene, which spans 290 kb on chromosome 21 and is actively transcribed. To avoid the impact of foreign gene expression on chromatin dynamics, we only inserted 8×ParSc 12 kilobases upstream (S3 locus marked as the promoter region) and 8×ParSm 12 kilobases downstream (S4 locus marked as the terminator region) of the APP gene (Fig 4A). We transfected TtParB-SNAP to visualize the S3 locus and TtParBm-HaloTag to visualize the S4 locus. As shown in Fig 4B, the S3 locus (8×ParSc) and S4 locus (8×ParSm) were visualized simultaneously in the U2OS-8×ParSc-APP-8×ParSm cells. We also test the influence of the ParSite system on gene transcription. The RT-qPCR result reveals that the ParSite system barely affect APP gene’s transcriptional activity (S6 Fig). As statistical analysis shown in Fig 4C, the 2D spatial distance between S3 (promoter region) and S4 (terminator region) ranges from 37 nm to 697 nm with 284 nm on average from 3D projection, which indicates the distance is heterogeneously distributed. Previous research shows that the terminator region will gradually reaches the promoter region, forming a chromatin contact “stripe” feature in the Hi-C map during transcription [34], suggesting the dynamic interaction between the gene’s promoter and terminator. As shown in Fig 4D, live cell tracking of the 2D distance from 3D projection between S3 and S4 over 40 s among several cells revealed that promoter and terminator regions of the APP gene maintained the spatial distances in each cell. A similar observation was seen when tracking for 8 min (Fig 4E and S1 Video).
Fig 4. Tracking the dynamics of the promoter and terminator of the APP gene by the ParSite system.
(A) Schematic of labeling promoter and terminator of the APP gene by ParSite. TtParB-SNAP and TtParBm-HaloTag were transfected together into cell line U2OS-8×ParSc-APP-8×ParSm to label the loci S3 (promoter region) and S4 (terminator region). PAPP is the promoter region of the APP gene and TAPP is the terminator region of the APP gene. (B) Representative images of S3 and S4 loci labeled by the ParSite system. The images of Cell 1 and Cell 2 are captured from the same single-cell derived U2OS-8×ParSc-APP-8×ParSm line. The diagrams on the right indicate the gene loop states. Scale bars, 5 μm for images with the whole cells and 1 μm for zoomed images. (C) Statistics of 2D spatial distance between S3 and S4. The red line indicates the mean. n = 32. (D) Live tracking the 2D distances between the promoter and terminator of the APP gene. The distances between the promoter (S3) and terminator (S4) of the APP gene at each time point over 40 s were measured and plotted. (E) Time-lapse of S3 and S4 loci pair. This pair of loci was tracked over 8 min with a time interval of 1 min. Scale bars, 1 μm. The underlying data associated with this figure are available in S4 Data.
A ParSc-ParSm hybrid system for non-repetitive DNA imaging
Based on the ParB-ParS crystal structures, ParB dimer binds the 16 bp palindromic ParS sequence. TtParB dimer or TtParBm dimer binds to 16 base-pair palindromic ParSc or ParSm without cross-reaction. Here, we test whether the hybrid (ParSh) of half-ParSc and half-ParSm could be specifically recognized by TtParB and TtParBm heterodimer (Fig 5A). We knocked in 8×ParSh at the S1 locus (36 kb downstream of the C3 repeat) in U2OS cells (S1A and S1B Fig). We transfected TtParB and/or TtParBm to visualize the S1 locus, along with dCas9-GFP/sgRNA-C3 for labeling C3 locus (Fig 5B). As shown in Fig 5C and 5D, the S1 locus (8×ParSh in magenta) was not labeled by the TtParB dimer or TtParBm dimer but efficiently labeled by the TtParB-TtParBm heterodimer. Live tracking the S1 locus containing 8×ParSh, we saw the signal foci of TtParBm-HaloTag overlapped with TtParB-SNAP (S7 Fig). The signal intensity from ParBm and ParB spreading around ParSh is positively correlated with their relative expression level (Fig 5E). We also combine the 8×ParSm and 8×ParSh to achieve dual-color genomic loci labeling (S8 Fig). The ParSc-ParSm hybrid data enable us, for the first time, to use ParS label genome DNA without a palindromic sequence. Therefore, 8×ParSc, 8×ParSm, and 8×ParSh could be utilized for triple-color DNA imaging, and we termed this system as tParSite (tricolor ParSite).
Fig 5. Generation of a hybrid ParB-ParS system for live-cell DNA imaging.
(A) Structures of a hybrid ParB-ParS system. The heterodimer of TtParB (purple) and TtParBm (red) recognizes a ParS hybrid (ParSh) with a half ParSc and a half ParSm. The organizations of TtParB and TtParBm are based on the structure of Caulobacter vibrioides ParB (PDB entry 6T1F). The model in the right panel is viewed after a 90° rotation around the vertical axis. The helices that interact with the DNA molecule are labeled as α6, α8, and α10. (B) Diagram of the hybrid ParB-ParS system for DNA imaging. The 8×ParSh is integrated into the S1 locus, which is 36 kb downstream of the C3 repeat (as an endogenous reference locus). The hypothesis is that the heterodimer of TtParB and TtParBm will bind ParSh and could be used for DNA imaging. (C) Representative images of U2OS-8×ParSh labeled by TtParB-TtParBm heterodimer. TtParB only group and TtParBm only group are as controls. Scale bars, 5 μm for images with the whole cells and 1 μm for zoomed images. (D) The percentage of U2OS-8×ParSh cells labeled by TtParB-TtParB, TtParBm-TtParBm, or TtParB-TtParBm dimmers, respectively. n = 62 cells. (E) The quantification of ParSh relative labeling efficiency by ParB and ParBm. n = 42. One data point is outside the axis limit. The underlying data associated with this figure are available in S5 Data.
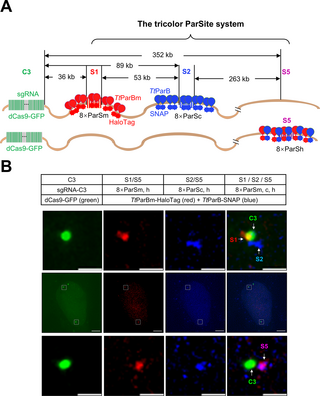
Simultaneously labeling 3 genomic loci by tricolor ParSite
To confirm that the tricolor ParSite (tParSite) could simultaneously label 3 target sites, we knocked in 8×ParSm at the S1 locus (36 kb downstream of the C3 locus), 8×ParSc at the S2 locus (89 kb downstream of the C3), and 8×ParSh at the S5 locus (352 kb downstream of the C3 repeat) into U2OS cells. To get the cell lines with knocked in 3 above 8×ParS derived sequences, we established a knock-in method with high integration efficiency resulting in 62% for 8×ParSm, 96.15% for 8×ParSc, and 100% for 8×ParSh (S1 Fig). We transfected TtParBm-HaloTag to visualize the S1 locus, TtParB-SNAP to visualize the S2 locus, TtParBm-HaloTag/TtParB-SNAP together to visualize the S5 locus, along with dCas9-GFP/sgRNA-C3 for labeling C3 repeat (Fig 6A). As shown in the top row of Fig 6B, C3 (green) and S1 (red) are partially overlapped, and both of them are proximal to S2 (blue). As shown in the bottom row of Fig 6B, C3 is proximal to S5 (purple was generated from red and blue overlapping). We also live-tracked the S1, S2, and S5 in the single cell (S2 and S3 Videos). These data suggested we generated a U2OS cell line with one allele containing 8×ParSm at S1 and 8×ParSc at S2 and another allele containing 8×ParSh at S5, which can achieve 3 color DNA imaging with ParSc, ParSm, ParSh, TtParB, and TtParBm. The labeling efficiency is 33.3% for tParSite (S4 Fig). It is easier to separate the 3 DNA loci of ParSc, ParSm, and ParSh if they located on different chromosomes than on one chromosome. Live cell tracking is needed if these 3 loci integrated into same chromosome, and the distance between a pair of them is critical for later spot analysis.
Fig 6. Labeling 3 genomic DNA loci simultaneously by tricolor ParSite.
(A) Simplified schematic of the tricolor ParSite system for simultaneous imaging of 3 genomic loci; 8×ParSm, 8×ParSc, and 8×ParSh were integrated into U2OS at the S1, S2, and S5 loci, respectively, downstream of C3 repeat on chromosome 3. The dimeric TtParBm-HaloTag, dimeric TtParB-SNAP, and heterodimeric TtParBm-HaloTag/TtParB-SNAP were used for labeling S1, S2, and S5, respectively. (B) Representative images of simultaneous imaging of 3 genomic loci in single cell. U2OS-8×ParSm-8×ParSc-8×ParSh cell line was transfected with TtParB-SNAP and TtParBm-HaloTag, along with dCas9-GFP and sgRNA-C3 for labeling of C3 repeats as a reference point. The middle panel was the whole cell nucleus. The upper panel shows the labeling of S1 (red) and S2 (blue) loci adjacent to C3 (green). The below panel shows the labeling of S5 (magenta) locus adjacent to C3. Scale bars, 5 μm for images with the whole cells and 1 μm for zoomed images.
Discussion
Genome organization in space and time is critical for transcriptional regulation, cell growth, and differentiation. There are extensive studies on genome organization in 3D in fixed cells by either sequencing such as Hi-C, or imaging such as DNA FISH [35]. However, the studies on genome organization in 4D (time is the fourth dimension) are lacking behind, mainly because of the challenge of imaging non-repetitive genomic loci in living cells. We previously developed mParSpot [30], a dual-color DNA imaging approach based on the ParB-ParS and Noc-NBS systems. The mParSpot allows to track pairwise genomic loci and explore genomic interactions. Here, we created ParSite, a multicolor DNA imaging approach by a two-step process: (1) rational design of the interface amino acids between TtParB and ParSc resulting in TtParBm and ParSm; (2) Generation of the ParSh with half-ParSc and half-ParSm allowing for non-repetitive genomic DNA imaging. Combining ParB-ParSc and ParBm-ParSm, we can realize dual color DNA imaging. Although we are unclear of the impact of ParSite on genome topology, ParSite hardly down-regulate gene transcription in our cell lines. Based on ParSc, ParSm, ParSh, ParB, and ParBm, we can achieve triple-color DNA imaging. Importantly, the tricolor ParSite will allow us to track the contour of multiple loci on chromosomes in living cells. This will be important to provide information on DNA loop dynamics at a single-cell level over time.
ParB typically spreads a few kilobases around ParS from Chip-seq in bacteria [36]. The number of ParB proteins around the ParS site for the DNA labeling in mammalian cells is worthwhile considering. To estimate the amount of ParB proteins around the integrated ParS site, we compared the intensity of TetR on 120×TetO with ParB around 8×ParS. It was estimated an average of 52 ParB or 30 ParBm proteins around the ParSc or ParSm site, respectively (S9 Fig). The spot size of ParB around the ParS site was measured to be less than 300 nm under the wide-field microscope we used. The spot size and shape could be more precise if we could use the super-resolution microscope for live cell DNA imaging by the ParSite system in future studies. The SNR is negatively correlated with the expression level of ParB, which indicates that background reduction could also enhance the sensitivity of DNA imaging by the ParSite system.
ParB and Noc are structurally similar, and the DNA-binding specificity of ParB to ParS can switch to NBS by mutations of 4 conserved amino acids [31]. However, it is unclear whether TtParB containing the above 4 mutations (TtParBm) can switch its binding specificity from ParSc to NBSc (ParSm), which is 4 nucleotides different in between, for DNA imaging in living cells. We found these mutations could successfully switch the binding specificity of TtParB for DNA imaging, although the SNRs decreased to some degree. To our surprise, the ParSh of half-ParSc and half-ParSm switches its binding specificity to the heterodimer TtParB and TtParBm, which led us to develop tricolor ParSite. These results, for the first time, demonstrated heterodimer ParB was able to recognize non-palindromic ParS hybrid and spread around the neighboring DNA. The detailed mechanism for the recognition and spreading of ParBs around the ParS site is unknown. We hypothesize that it might be the ParB-ParBm heterodimer recognition of the ParSh site and subsequently spreading around the same spot. Previously, we found the Noc-NBS system could be repurposed for DNA imaging [30]. To further expand the labeling colors for DNA loci, we developed the ParSite system to achieve triple-color DNA imaging. By adding one more color, we can not only measure the distance of 2 genomic loci over time but also potentially track the contour of 3 genomic locations in space and time. To better locate the 3 genomic spots labeled by tParSite, time-lapse tracking is useful for resolving the colocalization concern if they are close in genomic distance or spatial distance. To be concluded, the ParSite system is a simple and compact tool for DNA imaging in living cells with minimal disturbance in RNA transcription and cell growth.
It has been proposed that loop interactions among enhancers, promoters and terminators regulate the transcription of genes [34,37–41]. However, it is still not fully understood how the spatial distance and duration of promoter and enhancer (P-E) or promoter and terminator (P-T) interactions contribute to transcriptional activity in living cells. Here, we found that the spatial distance of APP’s promoter and terminator was maintained during the duration of 40 s, suggesting that the APP’s promoter and terminator loops are relatively stable in a short period. It will be intriguing to investigate whether the spatial distances and stability of promoter and terminator loops contribute to the transcriptional activity of the APP gene. Using tricolor ParSite (tParSite), we should be able to better define P-T loops with additional labeling of a locus in between the promoter and terminator. By combining with Cas13-based RNA imaging [42], we may investigate how the P-T loops contribute to transcriptional activity or bursts. In sum, the ParSite system is a useful addition to the DNA imaging toolbox and is complementary to the FROS system and CRISPR-based DNA imaging methods. With this strategy, introducing one more ParS independent of ParSc and ParSm could generate a color palette of ParB-ParS-based DNA imaging in living cells.
Methods
Plasmid construction
For single color genomic locus labeling, the donor plasmids used for the integration of 8×ParSc, 8×ParSm, and 8×ParSh consist of 4 regions: left homology arm, 8×ParSc, 8×ParSm, or 8×ParSh, puromycin resistant gene with CMV promoter and SV40 PolyA signal and right homology arm. For dual-color genomic loci tracking, the donor plasmids are similar as above, except that the hygromycin-resistant gene with EFS promoter was added as a second selection marker. For triple color DNA labeling, the bleomycin-resistant gene was used as a third selection marker. For gene promoter and terminator labeling, the donor plasmids have 4 regions: left homology arm, 8×ParSc or 8×ParSm, right homology arm, and inverse-oriented CMV promoters. All these donor plasmids were constructed from pDONOR3.1 and ligated with a seamless assembly enzyme mix. For TtParB or TtParBm expression vectors, pHAGE-EFS-MCP-HaloTag or pHAGE-EFS-N22P-SNAP was used for subcloning. The 53BP1 DNA fragments were codon-optimized and synthesized by Beijing Tsingke Biotech Co., Ltd. We synthesized a 30-mer TetO array from GenScript. The 120-mer TetO array was constructed with 4 copies of the 30-mer TetO array in a DONOR plasmid. Later, the CMV promoter and enhancer, Puromycin resistant gene, and SV40 PolyA fragments were integrated downstream of the 120-mer TetO array using seamless assembly. Finally, the left homology arm and right homology arm for DHFR or WEE1 are inserted into the 120-mer TetO plasmid, respectively. The core sequences of 8×ParS, 8×ParSm, 8×ParSh, and 120-mer TetO are listed in S1 Table. The sgRNA sequences are listed in S2 Table.
Generation of U2OS cell lines with 8×ParS integration
U2OS cells were cultured on 10 cm dishes at 37°C in DMEM with high glucose (Life Technologies) supplemented with 10% (vol/vol) FBS. To generate single 120-mer TetO and 8×ParS-integrated cell lines, U2OS cells were co-transfected with 1 μg of pHAGE-Cas9-P2A-sfGFP, 600 ng of pLB-sgRNA1 and 500 ng of pDONOR-8×ParSc, pDONOR-8×ParSm, pDONOR-8×ParSh, or pDONOR-120×TetO using Lipofectamine 2000 (Life Technologies) for 6 h and then replaced with fresh culture media. The transfected cells were cultured for an additional 48 h before flow cytometry to sort BFP and GFP double-positive cells by FACSAria III. The collected cells were plated on 48-well plates and cultured for an additional 24 h. Puromycin was added to enrich the cells with 8×ParSc, 8×ParSm, 8×ParSh, or 120×TetO integration for 7 days. Single cells were then sorted into 96-well plates and cultured for an additional 2 to 3 weeks. The single-cell clones were expanded and subjected to genotyping. To generate dual 8×ParS-integrated cells, the cell line was subjected for double selections with puromycin and hygromycin. To generate triple 8×ParS-integrated cells, the cells were subjected for triple selections with puromycin, followed by hygromycin and zeocin sequentially. Single-cell clones were identified with primers in S3 Table. We used 2 pairs of primers to identify whether the homozygous or heterozygous integration of 8×ParS into the cell lines.
Transfection for DNA imaging by the ParSite system
For 8×ParSc or 8×ParSm labeling test, 600 ng of SgRNA-C3 repeat, 300 ng of dCas9-GFP, and 500 ng of TtParB-HaloTag or TtParBm-HaloTag are transfected together into U2OS cells using Lipofectamine 2000 (Life Technologies). For 8×ParSh-mediated DNA imaging, 8×ParSc and 8×ParSm-mediated dual color DNA imaging or 8×ParSc, 8×ParSm, and 8×ParSh-mediated triple color DNA imaging, 600 ng of sgRNA-C3 repeat, 300 ng of dCas9-GFP and 600 ng of TtParB-SNAP and/or 500 ng of TtParBm-HaloTag are transfected together into cells.
DNA double-strand breaks detection in live cells by GFP-53BP1
We chose 53BP1 as a marker for DNA DSBs in live cells. The locus of downstream C3 repeat and/or upstream S2 were cleavage sites. The day before transfection, S1-ParSm-S2-ParSc-integrated cell lines were cultured into 35 mm glass bottom dish from 10 cm culture dish, and 400 ng of sgRNA-C3 repeat, 1.5 μg of SpCas9-BFP-U6-sgRNA1-U6-sgRNA2, 400 ng of TtParBm-HaloTag, 1 μg of TtParB-SNAP, and 300 ng of GFP-53BP1 were transfected together into S1-ParSm-S2-ParSc integrated cells using EZ Trans Transfection Reagent or Lipofectamine 2000 for 6 h, and then replaced with fresh culture media for additional 18 h. HaloTag-JF549 and SNAP-Cell 647-SiR were added into the dish 8 h before imaging. For the control group, 400 ng of sgRNA-C3 repeat, 331 ng of SpCas9-BFP, 400 ng of TtParBm-HaloTag, 1 μg of TtParB-SNAP, and 300 ng of GFP-53BP1 were transfected into S1-ParSm-S2-ParSc cells.
Tracking the promoter and terminator interactions of the APP gene by the ParSite system
For promoter and terminator labeling, 600 ng of TtParB-SNAP and 500 ng of TtParBm-HaloTag were transfected together into ParS-integrated cells using EZ Trans Transfection Reagent for 8 h and replaced with fresh culture media then for 24 h before imaging.
RT-qPCR
The U2OS-8×ParSc-APP-8×ParSm cells were cultured in a 6-cm dish, and 1.3 μg of TtParB-HaloTag or control plasmids were transfected into cells. One day after transfection, cells were collected to sort transfected cells by Arial III. Total RNA was extracted using the RNAprep Pure Micro Kit (Tiangen Biotech Co., Ltd.). We utilized ABScript III RT Master Mix for qPCR with gDNA Remover and 2X Universal SYBR Green Fast qPCR Mix (ABclonal) to do RT-qPCR reaction. The primer sequences are listed in S3 Table.
Triple color DNA imaging in live cells by tParSite system
Approximately 16 h after transfection, HaloTag-JF549 and SNAP-Cell 647-SiR were added to the dish. The image is acquired 24 h after transfection. To verify and separate the triple spots labeled by tParSite system (ParB/ParSc, ParBm/ParSm, ParB-ParBm/ParSh), we recommend testing the comovement of ParB and ParBm on each locus by live tracking. For ParSc and ParSm, the fluorescence signals of ParB and ParBm would separate most of the time or at a certain time during tracking. For ParSh, the fluorescence signals of ParB and ParBm would be most likely overlapped all the time during tracking.
Fluorescence microscopy
The live-cell DNA imaging was carried out on a DeltaVision Ultra imaging system equipped with a 100× PlanApo oil objective lens (NA 1.4). The cells were cultured on No. 1.0 glass bottom dishes (MatTek). BFP was excited with an excitation filter at 397/31 nm, and its emission was collected using an emission filter at 438/36 nm. sfGFP was excited at 478/28 nm and collected using the filter at 512/23 nm. HaloTag-JF549 was excited at 548/34 nm, and its emission was collected using the filter at 592/38 nm. SNAP-Cell 647-SiR was excited with an excitation filter at 633/27 nm, and its emission was collected using an emission filter at 677/46 nm. The fluorescence imaging data were acquired by DeltaVision Elite imaging software. The images were captured in z-stacks with an exposure time of 50 ms under 50% laser power for HaloTag, GFP, BFP, or SNAP, respectively. The step size in z-stacks was 200 nm. For the representative images, the raw data were deconvoluted and projected by softWoRx software.
Statistical analysis
The bar graph, dot plot, and broken line chart are produced by GraphPad Prism software. Experiments with representative data are conducted at least 3 times unless otherwise stated. Images are randomly captured in different fields of interest and processed one by one, excluding non-complete or worse pictures. Data are processed with Excel. The labeling efficiency is estimated from all transfected cells. The SNR was calculated with the formula: SNR = (IS-IB)/(IN-IB). IS is the intensity of the labeled loci; IN is the intensity of the nucleoplasm; and IB is the background fluorescence intensity from a dark region in the same image.
The zoom-in images are processed in ImageJ by adjusting the minimum and maximum fluorescence intensity range. The background fluorescence intensity of proteins is calculated with IN-IB (the IN and IB are mentioned above) and measured by ImageJ. The 2D distances are calculated with the formula in Excel. The position of the spot is measured by ImageJ. The spot area is marked with an irregular shape around its contour and measured by ImageJ. The 3D distances are calculated by Imaris with its spot module.
The percentage of cells grouping in Fig 3B–3D is calculated within cells containing 53BP1 foci, and the foci are close or overlap with C3 or S2 spots.
Relative SNR in Fig 5E is calculated with the formula: relative SNR = (IS-IB)ParBm/(IS-IB)ParB. The relative expression level of ParBm versus ParB is calculated with the formula: Relative expression levelParBm/ParB = (IN-IB)ParBm/(IN-IB)ParB. IS is the signal fluorescence intensity for ParB or ParBm at the ParSh site. IN is the nuclear intensity of ParBm or ParB expressed cells. IB is the fluorescence intensity of the dark region in the same image.
The mean intensity of the signal spot in S9 Fig is calculated with the formula: mean intensity = IS-IB. IS is the fluorescence intensity of signal foci, and IB is the fluorescence intensity of the dark region in the same image.
Supporting information
S1 Fig. Integration of 8×ParS adjacent to C3 repeat in the U2OS cells.
(A) The schematic of sequential integration of 8×ParSs into different genomic target sites. The 8×ParS-1was firstly integrated into a target site along with the expression cassette of selection marker puromycin. The 8×ParS-2 was integrated into another target site along with the expression cassette of selection marker hygromycin. The 8×ParS-3 was integrated into a third target site along with the expression cassette of selection marker zeocin. (B) Clonal selection by FACS. Donor plasmids containing 8×ParS-1 and puromycin resistant gene along with Cas9/sgRNA were transfected into U2OS cells. Puromycin was used to kill these cells without transfected and FACS was applied for the single-cell selection. The second donor plasmid contains 8×ParS-2 and hygromycin resistant gene and hygromycin was used to select 8×ParS-2 integrated cells. The third donor plasmid contains 8×ParS-3 and zeocin resistant gene. Zeocin was used to select 8×ParS-3 integrated cells. Through these steps to improve integration efficiency. (C) Integration efficiency of 8×ParS into U2OS cells. The integration efficiency was estimated by counting the positive clones with 8×ParSm, 8×ParSc, or 8×ParSh from passaged single-cell clone. The underlying data associated with this figure are available in S6 Data.
https://doi.org/10.1371/journal.pbio.3003009.s004
(TIF)
S2 Fig. The correlation of SNR and ParB expression.
(A) The quantification of SNR from ParB/ParS system by the expression level of ParB protein. n = 50. (B) The quantification of SNR from ParBm/ParSm by the expression level of ParBm protein. n = 46. The underlying data associated with this figure are available in S7 Data.
https://doi.org/10.1371/journal.pbio.3003009.s005
(TIF)
S3 Fig. The 3D distance and area of C3, S1, and S2 loci.
(A) The representative zoom-in images of C3 (green), S1 (brown), and S2 (red) spots. The left is the original processed image and the right is the spot-module mimic image. Scale bars, 0.2 μm. (B) The distribution of 3D distance with C3-S1, S1-S2, and C3-S2. Red lines indicate the mean value. n = 42. (C) The area measurement of C3, S1, and S2 spots. n = 37. The red line indicates the mean area of each group. The underlying data associated with this figure are available in S8 Data.
https://doi.org/10.1371/journal.pbio.3003009.s006
(TIF)
S4 Fig. The labeling efficiency of double- and triple-color DNA labeling.
(A) The diagram of double and triple color DNA labeling. (B) Labeling efficiency of double- and triple-color DNA imaging. n = 42 cells for double-label, n = 42 cells for triple-label. The underlying data associated with this figure are available in S9 Data.
https://doi.org/10.1371/journal.pbio.3003009.s007
(TIF)
S5 Fig. The representative images of dParSite cells when no cleavage is induced.
SgRNA-C3, SpCas9-BFP, GFP-53BP1, ParBm-HaloTag, and ParB-SNAP plasmids are transfected together into cells. Arrowheads indicate the positions of C3, S1, and S2 loci. Scale bars, 5 μm.
https://doi.org/10.1371/journal.pbio.3003009.s008
(PDF)
S6 Fig. Fold change of APP mRNA level in the control group and ParS/ParB group.
We transfect TtParB plasmid or control plasmid into U2OS-8×ParSc-APP-8×ParSm cells. Total RNA is extracted to complete RT-qPCR testing the expression level of APP gene. The underlying data associated with this figure are available in S10 Data.
https://doi.org/10.1371/journal.pbio.3003009.s009
(TIF)
S7 Fig. Live cell tracking of the S1 locus labeled by the ParB-ParS hybrid system.
(A) The diagram of labeling the S1 locus by the ParB-ParS hybrid system; 8×ParSh was integrated into the S1 locus and labeled by the heterodimer of TtParB and TtParBm. (B) Time-lapse of the S1 locus labeled by the ParB-ParS hybrid system. Both TtParBm-HaloTag and TtParB-SNAP labeled the S1 locus in the ParB-ParS hybrid system. The S1 locus was tracked over 56 s. Scale bars, 0.5 μm. (C) Plots of the fluorescence intensity of the S1 locus indicated by the dashed lines in B. Red line represents the fluorescence intensity of TtParBm-HaloTag. Blue line represents the fluorescence intensity of TtParB-SNAP. The length of each dashed line is 0.7 μm.
https://doi.org/10.1371/journal.pbio.3003009.s010
(TIF)
S8 Fig. Simultaneous imaging of 2 DNA loci by 8×ParSm and 8×ParSh.
(A) The schematic of labeling S1 and S5 loci by ParSm or ParSh; 8×ParSm and 8×ParSh were integrated into U2OS at the S1 and S5 loci respectively, downstream of C3 repeat on chromosome 3. The dimeric TtParBm-HaloTag and heterodimeric TtParBm-HaloTag/TtParB-SNAP were used for labeling S1 or S5, respectively. (B) Representative images of simultaneous imaging of the S1 and S5 loci. TtParB-SNAP and TtParBm-HaloTag along with dCas9-GFP and sgRNA-C3 for labeling of C3 repeats are transfected into cells to label S1, S5, and C3 repeat loci. The bottom panel shows the simultaneous labeling of C3 (green), S1 (red), and S5 (magenta). Scale bars, 5 μm for images with the whole cells and 1 μm for zoomed images.
https://doi.org/10.1371/journal.pbio.3003009.s011
(TIF)
S9 Fig. The intensity comparison of TetO/TetR DNA labeling system and ParB/ParS system.
(A) The labeling strategy for DHFR and WEE1 genes by 120-mer TetO array. (B) Representative images of DHFR and WEE1 genes labeling with TetO/TetR system. Scale bars, 5 μm. (C) The SNR of 120-mer TetO/TetR system. n = 29 for DHFR group, and n = 40 for WEE1 group. Red lines indicate mean value. (D) The mean intensity of signal foci for 120-mer TetODHFR/TetR, 120-mer TetOWEE1/TetR, 8-mer ParSc/ParB, and 8-mer ParSm/ParBm systems. n = 32, 39, 48, 46 for each group from left to right. Red line indicates mean value for each group. The underlying data associated with this figure are available in S11 Data.
https://doi.org/10.1371/journal.pbio.3003009.s012
(TIF)
S1 Video. Tracking the dynamics of promoter and terminator region of APP gene by ParSite.
The video is shown in a total time of 8 min. The interval between each picture is 1 min. Images were cropped to 50 × 50 pixels. Scale bar, 1 μm. The playback rate is 5 frames per second. Green: S3 locus (promoter region) and Red: S4 locus (terminator region).
https://doi.org/10.1371/journal.pbio.3003009.s013
(MP4)
S2 Video. Time-lapse tracking of S1 and S2 loci in tParSite cells.
The video scan time is 66 s. The scan sequence is first Channel then Z to minimize time delay effect. Images were cropped to 50 × 50 pixels. Scale bar, 1 μm. The playback rate is 10 frames per second. Blue: S2 locus, Green: C3 locus, Red: S1 locus.
https://doi.org/10.1371/journal.pbio.3003009.s014
(MP4)
S3 Video. Time-lapse tracking of S5 loci in tParSite cells.
The video scan time is 66 s. The scan sequence is first Channel then Z to minimize time delay effect. Images were cropped to 50 × 50 pixels. Scale bar, 1 μm. The playback rate is 10 frames per second. Green: C3 locus, magenta (blue + red): S5 locus.
https://doi.org/10.1371/journal.pbio.3003009.s015
(MP4)
Acknowledgments
We thank Luke Lavis (Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA) for the HaloTag JF-549. DeltaVision Ultra microscopy was provided by Shanghai Institute for Advanced Immunochemical Studies (SIAIS) at ShanghaiTech University and Fluorescence activated cell sorting (FACS) was provided by the School of Life Science and Technology and Shanghai Institute for Advanced Immunochemical Studies (SIAIS) at ShanghaiTech University. We thank Lu Zhuang and also the Molecular Imaging Core Facility at ShanghaiTech University for their help with Imaris software.
References
- 1.
Ghosh RP, Meyer BJ. Spatial Organization of Chromatin: Emergence of Chromatin Structure During Development. Annu Rev Cell Dev Biol. 2021;37:199–232. Epub 20210706. pmid:34228506. - 2.
Arnould C, Rocher V, Saur F, Bader AS, Muzzopappa F, Collins S, et al. Chromatin compartmentalization regulates the response to DNA damage. Nature. 2023;623(7985):183–192. pmid:37853125 - 3.
Song F, Chen P, Sun D, Wang M, Dong L, Liang D, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344(6182):376–380. pmid:24763583. - 4.
Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357(6349). pmid:28751582. - 5.
Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261(5118):203–206. pmid:8327891. - 6.
Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. pmid:11847345. - 7.
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. pmid:19815776. - 8.
Rao Suhas SP, Huntley Miriam H, Durand Neva C, Stamenova Elena K, Bochkov Ivan D, Robinson James T, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–1680. pmid:25497547 - 9.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. pmid:22495300 - 10.
Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci U S A. 2012;109(52):21301–6. Epub 20121211. pmid:23236188. - 11.
Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362(6413). pmid:30361340. - 12.
Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell. 2020;182(6):1641–59.e26. Epub 20200820. pmid:32822575. - 13.
Cardozo Gizzi AM, Cattoni DI, Fiche J-B, Espinola SM, Gurgo J, Messina O, et al. Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol Cell. 2019;74(1):212–22.e5. pmid:30795893 - 14.
Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019;568(7750):49–54. pmid:30886393 - 15.
Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135(6 Pt 2):1685–1700. pmid:8991083. - 16.
Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. pmid:9335333. - 17.
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491. pmid:24360272. - 18.
Chen B, Hu J, Almeida R, Liu H, Balakrishnan S, Covill-Cooke C, et al. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016;44(8):e75. Epub 20160105. pmid:26740581. - 19.
Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015;112(10):3002–7. Epub 20150223. pmid:25713381. - 20.
Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol. 2016;34(5):528–30. Epub 20160418. pmid:27088723. - 21.
Ma H, Tu LC, Naseri A, Chung YC, Grunwald D, Zhang S, et al. CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging. Nat Methods. 2018;15(11):928–31. Epub 20181030. pmid:30377374. - 22.
Saad H, Gallardo F, Dalvai M, Tanguy-le-Gac N, Lane D, Bystricky K. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 2014;10(3):e1004187. Epub 20140313. pmid:24625580. - 23.
Germier T, Kocanova S, Walther N, Bancaud A, Shaban HA, Sellou H, et al. Real-Time Imaging of a Single Gene Reveals Transcription-Initiated Local Confinement. Biophys J. 2017;113(7):1383–1394. pmid:28978433. - 24.
Germier T, Audibert S, Kocanova S, Lane D, Bystricky K. Real-time imaging of specific genomic loci in eukaryotic cells using the ANCHOR DNA labelling system. Methods. 2018;142:16–23. Epub 20180413. pmid:29660486. - 25.
Delker RK, Munce RH, Hu M, Mann RS. Fluorescent labeling of genomic loci in Drosophila imaginal discs with heterologous DNA-binding proteins. Cell Rep Methods. 2022;2(3):100175. Epub 20220309. pmid:35475221. - 26.
Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, et al. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. Elife. 2019;8. Epub 20190524. pmid:31124784. - 27.
Li J, Dong A, Saydaminova K, Chang H, Wang G, Ochiai H, et al. Single-Molecule Nanoscopy Elucidates RNA Polymerase II Transcription at Single Genes in Live Cells. Cell. 2019;178(2):491–506.e28. Epub 20190530. pmid:31155237. - 28.
Lyu XY, Deng Y, Huang XY, Li ZZ, Fang GQ, Yang D, et al. CRISPR FISHer enables high-sensitivity imaging of nonrepetitive DNA in living cells through phase separation-mediated signal amplification. Cell Res. 2022;32(11):969–81. Epub 20220914. pmid:36104507. - 29.
Guilhas B, Walter JC, Rech J, David G, Walliser NO, Palmeri J, et al. ATP-Driven Separation of Liquid Phase Condensates in Bacteria. Mol Cell. 2020;79(2):293–303.e4. pmid:32679076. - 30.
He X, Tan Y, Feng Y, Sun Y, Ma H. Tracking pairwise genomic loci by the ParB-ParS and Noc-NBS systems in living cells. Nucleic Acids Res. 2024;52(9):4922–4934. pmid:38412314. - 31.
Jalal ASB, Tran NT, Stevenson CE, Chan EW, Lo R, Tan X, et al. Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family. Cell Rep. 2020;32(3):107928. pmid:32698006. - 32.
Wang H, Nakamura M, Abbott TR, Zhao D, Luo K, Yu C, et al. CRISPR-mediated live imaging of genome editing and transcription. Science. 2019;365(6459):1301–5. Epub 20190905. pmid:31488703. - 33.
Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, et al. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J Cell Biol. 2016;214(5):529–37. Epub 20160822. pmid:27551060. - 34.
Zheng M, Tian SZ, Capurso D, Kim M, Maurya R, Lee B, et al. Multiplex chromatin interactions with single-molecule precision. Nature. 2019;566(7745):558–562. pmid:30778195 - 35.
Kempfer R, Pombo A. Methods for mapping 3D chromosome architecture. Nat Rev Genet. 2020;21(4):207–26. Epub 20191217. pmid:31848476. - 36.
Osorio-Valeriano M, Altegoer F, Das CK, Steinchen W, Panis G, Connolley L, et al. The CTPase activity of ParB determines the size and dynamics of prokaryotic DNA partition complexes. Mol Cell. 2021;81(19):3992–4007.e10. Epub 20210924. pmid:34562373. - 37.
Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. pmid:22682246. - 38.
Schoenfelder S, Fraser P. Long-range enhancer–promoter contacts in gene expression control. Nat Rev Genet. 2019;20(8):437–455. pmid:31086298 - 39.
Bartman Caroline R, Hsu Sarah C, Hsiung Chris CS, Raj A, Blobel GA. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol Cell. 2016;62(2):237–247. pmid:27067601 - 40.
Kawasaki K, Fukaya T. Regulatory landscape of enhancer-mediated transcriptional activation. Trends Cell Biol. 2024;34(10):826–837. pmid:38355349 - 41.
Popay TM, Dixon JR. Coming full circle: On the origin and evolution of the looping model for enhancer-promoter communication. J Biol Chem. 2022;298(8). pmid:35691341 - 42.
Yang LZ, Wang Y, Li SQ, Yao RW, Luan PF, Wu H, et al. Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems. Mol Cell. 2019;76(6):981–97.e7. Epub 20191119. pmid:31757757.
ADVERTISEMENT:
Hai, para pencinta slots Pernah denger istilah “slot demo”? jika tidak, bersiaplah jatuh hati sama program ini. slot gacor adalah mesin slots yang selalu memberi win. Ya, slot-slot ini bisa dibilang sebagai andalannya buat bawa come back hasil. any way, cemana sih
tekniknya jumpain slot demo yang tepat? Tenang Bro, kita bahas tenang saja di tempat ini
Gaming tergaco saat sekarang hanya satu di Indonesia yaitu akan menyediakan return on Investment tertinggi
SEGERA hanya di :
Informasi mengenai KING SLOT, Segera Daftar Bersama king selot terbaik dan terpercaya no satu di Indonesia. Boleh mendaftar melalui sini king slot serta memberikan hasil kembali yang paling tinggi saat sekarang ini hanyalah KING SLOT atau Raja slot paling gacor, gilak dan gaco saat sekarang di Indonesia melalui program return tinggi di kingselot serta pg king slot
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
situs slot terbaru
situs slot terbaru permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
slot terbaru
slot terbaru permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
suara88 permainan paling top dan garansi imbal balik hasil besar bersama suara88.biz
sumo7777 permainan paling top dan garansi imbal balik hasil besar bersama sumo7777.com
supermoney888 permainan paling top dan garansi imbal balik hasil besar bersama supermoney888.biz
teratai88 permainan paling top dan garansi imbal balik hasil besar bersama teratai88.biz
thor88 permainan paling top dan garansi imbal balik hasil besar bersama thor88.biz
togelhk88 permainan paling top dan garansi imbal balik hasil besar bersama togelhk88.net
topjitu88 permainan paling top dan garansi imbal balik hasil besar bersama topjitu88.net
totosloto88 permainan paling top dan garansi imbal balik hasil besar bersama totosloto88.com
trisula888 permainan paling top dan garansi imbal balik hasil besar bersama trisula888.biz
udangbet88 permainan paling top dan garansi imbal balik hasil besar bersama udangbet88.net
via88 permainan paling top dan garansi imbal balik hasil besar bersama via88.biz
virusjp88 permainan paling top dan garansi imbal balik hasil besar bersama virusjp88.net
warga888 permainan paling top dan garansi imbal balik hasil besar bersama warga888.biz
waw88 permainan paling top dan garansi imbal balik hasil besar bersama waw88.biz
winjitu88 permainan paling top dan garansi imbal balik hasil besar bersama winjitu88.net
wisdom88 permainan paling top dan garansi imbal balik hasil besar bersama wisdom88.biz
wnitogel88 permainan paling top dan garansi imbal balik hasil besar bersama wnitogel88.com
yoyo888 permainan paling top dan garansi imbal balik hasil besar bersama yoyo888.biz
validtoto88 permainan paling top dan garansi imbal balik hasil besar bersama validtoto88.com
sule999 permainan paling top dan garansi imbal balik hasil besar bersama sule999.com
sule88 permainan paling top dan garansi imbal balik hasil besar bersama sule88.org
ss888bet permainan paling top dan garansi imbal balik hasil besar bersama ss888bet.com
sia77 permainan paling top dan garansi imbal balik hasil besar bersama sia77.info
seluang88 permainan paling top dan garansi imbal balik hasil besar bersama seluang88.com
satu88 permainan paling top dan garansi imbal balik hasil besar bersama satu88.biz
satu777 permainan paling top dan garansi imbal balik hasil besar bersama satu777.asia
rp88 permainan paling top dan garansi imbal balik hasil besar bersama rp88.biz
rp88 permainan paling top dan garansi imbal balik hasil besar bersama rp88.asia
rp88 permainan paling top dan garansi imbal balik hasil besar bersama rp77.live
qiuqiu88 permainan paling top dan garansi imbal balik hasil besar bersama qiuqiu88.biz
pt88 permainan paling top dan garansi imbal balik hasil besar bersama pt88.org
pt77 permainan paling top dan garansi imbal balik hasil besar bersama pt77.info
produk88 permainan paling top dan garansi imbal balik hasil besar bersama produk88.asia
mt88 permainan paling top dan garansi imbal balik hasil besar bersama mt88.org
mt77 permainan paling top dan garansi imbal balik hasil besar bersama mt77.biz
menang66 permainan paling top dan garansi imbal balik hasil besar bersama menang66.biz
latobet888 permainan paling top dan garansi imbal balik hasil besar bersama latobet888.org
kedai96 permainan paling top dan garansi imbal balik hasil besar bersama kedai96.org
kedai188 permainan paling top dan garansi imbal balik hasil besar bersama kedai188.biz
ids88 permainan paling top dan garansi imbal balik hasil besar bersama ids88.biz
hp88 permainan paling top dan garansi imbal balik hasil besar bersama hp88.org
hp77 permainan paling top dan garansi imbal balik hasil besar bersama hp77.org
gm88 permainan paling top dan garansi imbal balik hasil besar bersama gm88.asia
gm77 permainan paling top dan garansi imbal balik hasil besar bersama gm77.net
final888 permainan paling top dan garansi imbal balik hasil besar bersama final888.org
duit88 permainan paling top dan garansi imbal balik hasil besar bersama duit88.asia
duit168 permainan paling top dan garansi imbal balik hasil besar bersama duit168.biz
divisi88 permainan paling top dan garansi imbal balik hasil besar bersama divisi88.org
dewi500 permainan paling top dan garansi imbal balik hasil besar bersama dewi500.biz
devil88 permainan paling top dan garansi imbal balik hasil besar bersama devil88.info
cuputoto88 permainan paling top dan garansi imbal balik hasil besar bersama cuputoto88.com
cukongbet88 permainan paling top dan garansi imbal balik hasil besar bersama cukongbet88.asia
bom888 permainan paling top dan garansi imbal balik hasil besar bersama bom888.biz
bintaro888 permainan paling top dan garansi imbal balik hasil besar bersama bintaro888.info
askasino88 permainan paling top dan garansi imbal balik hasil besar bersama askasino88.org
999aset permainan paling top dan garansi imbal balik hasil besar bersama 999aset.com
afb77 permainan paling top dan garansi imbal balik hasil besar bersama afb77.biz
aset99 permainan paling top dan garansi imbal balik hasil besar bersama aset99.biz
bendera77 permainan paling top dan garansi imbal balik hasil besar bersama bendera77.biz
bendera888 permainan paling top dan garansi imbal balik hasil besar bersama bendera888.com
coco88 permainan paling top dan garansi imbal balik hasil besar bersama coco88.org
cuma77 permainan paling top dan garansi imbal balik hasil besar bersama cuma77.biz
cuma88 permainan paling top dan garansi imbal balik hasil besar bersama cuma88.org
dwv88 permainan paling top dan garansi imbal balik hasil besar bersama dwv88.org
fafajp88 permainan paling top dan garansi imbal balik hasil besar bersama fafajp88.com
gemar88 permainan paling top dan garansi imbal balik hasil besar bersama gemar88.biz
gocap88 permainan paling top dan garansi imbal balik hasil besar bersama gocap88.info
gocaptoto permainan paling top dan garansi imbal balik hasil besar bersama gocaptoto.asia
hakabet88 permainan paling top dan garansi imbal balik hasil besar bersama hakabet88.com
hwtoto88 permainan paling top dan garansi imbal balik hasil besar bersama hwtoto88.org
ina77 permainan paling top dan garansi imbal balik hasil besar bersama ina77.biz
ina88 permainan paling top dan garansi imbal balik hasil besar bersama ina88.info
jingga8888 permainan paling top dan garansi imbal balik hasil besar bersama jingga8888.com
juragan777 permainan paling top dan garansi imbal balik hasil besar bersama juragan777.asia
kastil77 permainan paling top dan garansi imbal balik hasil besar bersama kastil77.info
kebo888 permainan paling top dan garansi imbal balik hasil besar bersama kebo888.biz
kkwin77 permainan paling top dan garansi imbal balik hasil besar bersama kkwin77.com
kokoslot88 permainan paling top dan garansi imbal balik hasil besar bersama kokoslot88.asia
luckydf88 permainan paling top dan garansi imbal balik hasil besar bersama luckydf88.org
microstar888 permainan paling top dan garansi imbal balik hasil besar bersama microstar888.biz
monperatoto88 permainan paling top dan garansi imbal balik hasil besar bersama monperatoto88.com
mpo1122 permainan paling top dan garansi imbal balik hasil besar bersama mpo1122.biz
mpo122 permainan paling top dan garansi imbal balik hasil besar bersama mpo122.biz
mpopelangi88 permainan paling top dan garansi imbal balik hasil besar bersama mpopelangi88.com
pamanslot88 permainan paling top dan garansi imbal balik hasil besar bersama pamanslot88.biz
panel88 permainan paling top dan garansi imbal balik hasil besar bersama panel88.org
paragon77 permainan paling top dan garansi imbal balik hasil besar bersama paragon77.biz
paragon888 permainan paling top dan garansi imbal balik hasil besar bersama paragon888.info
pion77 permainan paling top dan garansi imbal balik hasil besar bersama pion77.biz
prada88 permainan paling top dan garansi imbal balik hasil besar bersama prada88.asia
prada888 permainan paling top dan garansi imbal balik hasil besar bersama prada888.com
qqslot88slot permainan paling top dan garansi imbal balik hasil besar bersama qqslot88slot.com
rejekibet88 permainan paling top dan garansi imbal balik hasil besar bersama rejekibet88.com
rezekibet88 permainan paling top dan garansi imbal balik hasil besar bersama rezekibet88.org
sensa77 permainan paling top dan garansi imbal balik hasil besar bersama sensa77.biz
sensa888 permainan paling top dan garansi imbal balik hasil besar bersama sensa888.biz
singajp88 permainan paling top dan garansi imbal balik hasil besar bersama singajp88.com
sr77 permainan paling top dan garansi imbal balik hasil besar bersama sr77.org
sr88 permainan paling top dan garansi imbal balik hasil besar bersama sr88.org
surya77 permainan paling top dan garansi imbal balik hasil besar bersama surya77.biz
surya88 permainan paling top dan garansi imbal balik hasil besar bersama surya88.asia
tajir77 permainan paling top dan garansi imbal balik hasil besar bersama tajir77.info
tajir88 permainan paling top dan garansi imbal balik hasil besar bersama tajir88.biz
toto122 permainan paling top dan garansi imbal balik hasil besar bersama toto122.com
toto123 permainan paling top dan garansi imbal balik hasil besar bersama toto123.biz
uangvip88 permainan paling top dan garansi imbal balik hasil besar bersama uangvip88.com
wajik77 permainan paling top dan garansi imbal balik hasil besar bersama wajik77.asia
777neko permainan paling top dan garansi imbal balik hasil besar bersama 777neko.org
88judi permainan paling top dan garansi imbal balik hasil besar bersama 88judi.net
99judi permainan paling top dan garansi imbal balik hasil besar bersama 99judi.org
abcslot88 permainan paling top dan garansi imbal balik hasil besar bersama abcslot88.asia