The PAPCLEAR clinical study
The PAPCLEAR study is a monocentric cohort study that was designed to follow N = 150 women in the context of HPV infections (see ref. [28] for details about its protocole). Its inclusion criteria were to be between 18 and 25 years old, to live in the area of Montpellier (France), to be in good health (no chronic disease), not to have a history of HPV infection (e.g., genital warts or high-grade cervical lesion), and to report at least one new sexual partner over the last 12 months. Enrolment was independent of HPV infection and vaccination status. Given the lack of prior knowledge of HPV infections, the cohort size was chosen to maximise the number of inclusions and follow at least 75 infected women until infection clearance.
Participants were enrolled by putting up posters and handing out leaflets at the main STI detection centre (CeGIDD) within the University Hospital (CHU) of Montpellier (France) and in the Universities of the city. To increase enrolment, posters were also hung at bus stops near the CHU.
The inclusion visit (V1) was performed by a gynaecologist or a midwife at the CeGIDD outside operating hours. After an interview, biological samples were collected by the clinician during a gynaecological consult, after which a nurse collected blood samples and participants filled in a detailed questionnaire.
The second visit (V2) was scheduled four weeks later to inform the participants of the result of a screening test for cervical lesions (see also ref. [46]). Depending on their HPV status at the inclusion visit (see below), participants were oriented to the HPV–negative or to the HPV–positive arm of the study.
Participants in the HPV–negative arm, for whom no alphapapillomavirus was detected at V1, came back for on-site visits every four months. If an HPV was detected at one of the following visits, they switched to the HPV–positive arm. HPV–negative participants not infected by month 32 of the study were not followed anymore.
Participants in the HPV–positive arm had on-site visits every two months until clearance or chronification of the infection. Chronicity was defined as 24 months of infection by the same HPV genotype. Clearance of a specific infection was defined as two successive visits without any detection of the focal genotype.
The longitudinal cohort was complemented by a cross-sectional cohort, which was originally designed to enrol the same number of women, i.e., 150, who would only perform the inclusion visit (V1) and the results visit (V2). The inclusions began in November 2016 and, due to the COVID-19 pandemic and the associated strain on hospital services, the study was prematurely ended in September 2020. This led to the loss of follow-up of two participants and 110 non-inclusion in the cross-sectional cohort. Overall, we followed N = 189 women, including 149 (78.8%) in the longitudinal cohort, with a total of 974 on-site visits with gynaecological consults (Fig O in S1 Supplementary Materials).
Clinical samples description and processing
Cervical smears.
A cervical smear was collected at each of the on-site visits. The gynaecologist or midwife performed 2.5 turns before putting the brush into 20 mL Thinprep PreserCyt medium from Hologic (at V1) or 5 mL of fresh phosphate-buffered saline (PBS) medium (at the other visits).
Smears in PreserCyt medium were analysed by a trained pathologist of the CHU of Montpellier. Before that, 2 mL were sampled for HPV detection. Leftovers from the cytological analysis were also stored.
Smears in PBS were processed following a general protocol described in ref. [25]. The whole protocol was performed at 4°C. Within 2 h of collection, the smears were vortexed during 45 s with the cytobrush, which was then removed carefully to save as many cells as possible before adding 5 mL of Roswell Park Memorial Institute (RPMI) medium. The solution was then filtered in a new tube with a cell strainer (Fisherbrand Sterile Cell Strainers, 100 μm); 250 μL of the resulting solution were aliquoted for backup. The remaining solution was centrifuged for 10 min at 514 g. The resulting supernatant was stored at −80°C and the cell pellet was washed with 5 mL PBS at 514 g for 10 min. The supernatant was discarded, and cells were resuspended in 200 μL, and 20 μL of the resulting solution was aliquoted for HPV detection, while the rest was processed for flow cytometry staining.
Ophtalmic sponges.
As described in ref. [21], during the on-site visits, cervicovaginal secretions were collected by the gynaecologist or midwife using WeckCel sponges (Beaver-Visitec International) placed directly into the cervical os for 1 min. Sponges were then transferred into a Salivette (Sarstedt) device and centrifugated at 1,500 rpm for 5 min at 4°C after the addition of 175 μL of PBS. Supernatants were separated into 50 μL aliquots and stored at −80°C.
HPV detection and genotyping
HPV detection and typing was performed on the sample originating from the cervical smear and resuspended in 200 μL of PBS as described above. From this, we extracted DNA using the QIAamp DNA mini kit (QIAGEN Inc.) following standard protocol for body fluids (spin control).
We first tested for the presence of alphapapillomaviruses using the generic DEIA test [56] from DDL Diagnostic Laboratory (Rijswijk, the Netherlands). We then used the LiPA25 typing [57], also from DDL Diagnostic Laboratory, on DEIA-positive samples. Both assays target the same amplicon of the L1 HPV gene.
Samples that were DEIA-positive and LiPA25 negative were amplified using the PGMY PCR [58] and sequenced using Sanger sequencing. Samples for which the sequencing did not yield a clear sequence, most likely because of coinfections, were labelled as “non-typable.”
Genotype-specific HPV qPCR
We quantified the number of genomic copies by qPCR for 12 HPV genotypes (HPV16, 31, 35, 39, 51, 52, 53, 56, 58, 59, 66, and 68) using the protocols and primers from ref. [35]. We also quantified the number of copies of HPV18 and of a human reference gene (albumin). The HPV18 forward primer was 5′-ACACCACAATACCATGGCG-3′, the HPV18 reverse primer was 5′-TTCAGTTCCGTGCACAGATC-3′, and the HPV18 probe was 5′-FAM-CAACACG\GCGACCCTACAAGCTAC-HBQ1-3′. For the albumin primers, the details can be found in ref. [59].
All qPCRs were performed using SensiFAST Probe No-ROX Kit (MERIDIAN Bioscience) on LightCycler 96 and LightCycler 480 (Roche Diagnostics) for 384 wells plates. Cycles used for all genotypes and albumin were 95°C for 5 min and 40 two-step cycles with 95°C for 10 s and 60°C for 30 s. Each sample was run in triplicate with 2 μL of samples in a final volume of 10 μl. To create HPV standards, we used plasmids graciously provided by the international HPV reference centre from the Karolinska Instituet (https://www.hpvcenter.se). Each plasmid contained a full HPV genome for one of the 13 target genotypes. For the albumin standard, we used genomic female Human DNA (Promega). For each participant, we ran on the sample plate the qPCR for all samples, all genotypes previously identified by LiPa25 and the albumin gene.
Virus load measures were taken in triplicates and we analysed the mean cycle threshold (Ct) value. HPV genotype-specific Ct values were normalised by twice that of the cellular gene, the albumin (known to be present at two copies per diploid genome).
Cytokines quantification
We used the MSD U-plex Biomarker Group 1 (human) from Meso Scale Discovery (MSD, Rockville, Maryland, USA) to measure the concentration of five analytes identified in a previous study as being associated with HPV infection [21]: IFN-γ, IL-10, CCL20, IL-17A, and CXCL10. According to the manufacturer’s instructions, we used 25 μL of specimen, which was obtained from the ophtalmic sponges as detailed above. Analyses were performed on a MESO QuickPlex SQ 120 reader.
To normalise cytokine quantitation, we measured total protein concentration in the same samples using the Invitrogen Qubit protein assay (Thermo Fisher Scientific) following the manufacturer’s instructions.
To compute cytokine sample concentrations from the standard curves, we used a four-parameter logistic regression model implemented in the minpack.lm R package.
Antibody dosage
IgG and IgM antibodies targeting L1 proteins of high-risk HPV types 16, 18, 31, 33, 35, 45, 52, and 58, as well as low-risk HPV-types 6 and 11 were analysed with a multiplex serology assay using beads coated with recombinant glutathione s-transferase (GST) fusions proteins. The assay procedure has been previously described in detail [60]. The serum samples were tested at a final dilution of 1:100 using an IgG and an IgM goat anti-Human secondary antibody. Seropositivity was defined based on standard definitions [61,62]. We chose to focus on circulating antibodies in the serum rather than on mucosal antibodies in vaginal secretions because the latter are less abundant, they originate from samples that are difficult to standardise, and their variability is still poorly understood [63].
Flow cytometry
For the FCM analyses, within 3 h upon sampling, cellular suspensions were labelled with Live Dead red diluted 1/2,000 (Thermo Fisher) for 20 min in the dark, on ice. Cells were then washed with 2 mL of PBS (1,500 RPM, 5 min) and resuspended in 100 μL of PBS, before being transferred to a new tube containing dried antibodies (DURAClone, Beckman Coulter). No blocking step was undertaken before the labelling step. This custom antibody panel included anti-CD45 KRO (clone J33, IM2473U), anti-CD16 FITC (clone 3G8, B49215), anti-CD3 APC A750 (clone UCHT1, A94680), anti-CD4 APC A700 (clone 13B8.2, B10824), anti-CD8 PB (clone B9.11, A82791), anti-TCRγδPC5.5 (clone IMMU510, A99021), anti-CD69 PE (clone TP1.55.3, IM1943U), and anti-CD161 PC7 (clone 191B8, B30631).
Cells were incubated in the presence of antibodies for 15 min in the dark at room temperature, then washed with 2 mL of PBS (1,500 RPM, 5 min). Finally, cells were re-suspended in 250 μl of PBS 1% PFA and stored at 4°C until acquisition using a flow cytometer (Navios, Beckman Coulter).
From 631 acquired samples, 161 (25.5%) displayed visible pellets of red blood cells and were not analysed further.
Unsupervised FCM analysis
For unsupervised analyses, raw LMD files were converted to FCS 3.0 using a custom R script. Samples with visible blood pellets were excluded from the analysis.
Raw files were cleaned using the PeacoQC R package, and then filtered for viable cells (Lived Dead Red–). Among the 470 samples without visible red blood cells (RBC) pellets, 87 (17.7%) did not pass this quality control step and we eventually analysed 362 samples.
Using the CATALYST pipeline and FlowSOM, which builds on a Self-Organizing Map (SOM), an unsupervised technique for clustering and dimensionality reduction, we first restricted the data set to CD45+ cells [64,65].
We then clustered the cell populations using FlowSOM with 100 clusters and 20 metaclusters. The clustering was defined on all available samples, including both HPV–negative and HPV–positive samples. The following parameters were used to perform the clustering: SSC-A, FSC-A, CD45, CD3, CD4, CD8, TCRγδ and CD16. CD69 and CD161 were only used for differential expression analysis. The 20 metaclusters thus generated were manually merged into 11 cell populations (see Figs A and B and Table B in S1 Supplementary Materials).
A UMAP restricted to 150 cells per sample and on the same parameters used with FlowSoM clustering was performed to obtain a visual representation of the 11 cell population clusters.
We tested for differential abundances of the 11 cell populations between HPV–negative and focal HPV–positive samples using the diffcyt R package with the diffcyt-DA-edgeR method and including a random effect at the participant level. Differential expressions were calculated in a similar way using the diffcyt-DS-limma method. In both settings, adjusted p-values were obtained by applying a Benjamini–Hochberg procedure to correct for false discovery rate (FDR).
Basic statistical analyses
All statistical analyses were run with R version 4.4.
In Table 1 and Table A in S1 Supplementary Materials, we used respectively Kruskal–Wallis rank sum test kruskal.test function for numeric variables and the chisq.test function for the binary and categorical variables from the TableOne R package.
In Fig 2A, we used the brms R package to build a multivariate model. The five cytokines or chemokines were considered as separate response variables. Each participant was assumed to have a varying intercept. We assumed correlations between varying effects. The only fixed effect was the HPV focal status. The multivariate model was implemented with the mvbind function brms. To estimate the bayesian R2 in Table G in S1 Supplementary Materials, we used the loo_R2 function from the brms package.
In Fig 1B, we calculated correlation matrices with the lmer function (using the lme4 R package). Each cell population was considered as a response variable, with all five cytokines and chemokines as fixed effects, yielding 11 independent linear models. A random effect for the participant was also included in each model. The regression coefficient of each model is named “β” in the text.
Bayesian hierarchical modelling
All kinetics were modelled using rstan v. 2.32.3 (Stan version 2.26.1). The script used and raw data are available on https://doi.org/10.57745/KJGOYZ. We used the settings described in S1 Supplementary Materials in Table K for our models. The credible intervals were computed using the 2.5th and the 97.5th percentiles of the posterior distributions, i.e., with equally tailed intervals. In the figures, we systematically summarised the results by showing the median, interquartiles, and the 95% posterior distribution.
In the following, we describe the statistical modelling approach performed to infer the viral dynamics and the associated immune dynamics. To maximise the simplicity of the notations, represents the normal distribution with mean μ and standard deviation
, the multivariate normal distribution with variance-covariance matrix Σ, and ℰ(λ) represents the exponential distribution with rate λ. Finally, we use the notation v[n] to represent a vector v of length n.
Virus load dynamics.
qPCR data. This analysis included all the participants who had at least three on-site visits and at least one positive virus load with a genotype-specific qPCR (see Fig O in S1 Supplementary Materials).
For some participants, for a given genotype, there were HPV–negative visits between HPV–positive visits. In the analysis, we assumed that HPV–positive visits separated by two or more consecutive negative visits (i.e., at least four months) corresponded to two different infection events. Overall, this generated ten additional infections.
The qPCR data was pre-processed using the following steps:
- Samples with fewer than 100 copies of albumin and with HPV copies below the limit of detection (l.o.d.), i.e., three copies, were excluded;
- Samples with HPV copies below the l.o.d. and at least 100 copies of albumin were treated as censored at the limit of quantification (l.o.q.), i.e., ten copies;
- Samples with at least 100 albumin copies and HPV copies between the l.o.d. and the l.o.q. were considered interval-censored between one and ten copies;
- Samples with fewer than 100 albumin copies and HPV copies above the l.o.d. were deemed unreliable, with the number of HPV copies per cell considered above 10−4 (following the standard limit of detection [40]); and
- Samples with at least 100 copies of albumin and HPV copies above the l.o.q. were retained as is. The number of HPV copies per cell was then computed as the number of HPV copies divided by half the number of albumin copies.
The model.
Because of the shape of the raw data and of previous mechanistic mathematical modelling of HPV genital infections [29], we assumed that virus load dynamics followed a pattern with an exponential growth phase, a plateau, and a rapid clearance phase. These dynamics were captured using five parameters (Fig 4A) describing the log10 virus load, i.e., the number of HPV copies per cell, over time v(t), with t the time since the beginning of the follow-up. Formally, our virus load dynamics can be written as follows:
(1)
where, as illustrated in Fig 4A, the parameters indicate the plateau virus load (ψvl), the date of infection (ψt0), the growth phase duration (ψgr), the plateau duration (ψp), and the clearance phase duration (ψcl). v0 is the minimal viral load. All duration parameters are expressed per month, and ψvl and v0 are expressed as log10 (HPV copies/cell).
Estimation of the infection date.
To facilitate model convergence, we computed the starting date of each infection (ψt0) differently depending on its follow-up censoring status. For each participant i, each genotype g, and each infection j:
- When the follow-up was right-censored (i.e., the participant was not infected at inclusion but still infected at the last visit), we estimated ψt0 relatively to the observed infection time
(middle of last negative and first positive sample), assuming the informative prior
.
- When the follow-up was left-censored (i.e., the participant was infected at inclusion but at least one negative observation in the end), we estimated ψt0 relatively to the observed clearance time
(middle of the last positive and the first negative sample), assuming the informative prior
.
- When the follow-up was complete (i.e., at least one negative observation at inclusion and one negative observation in the end), we estimated ψt0 relatively to the observed midpoint of infection
, i.e., the mean date of all the genotype-specific positive samples from an individual. We the assumed the informative prior
.
- When the follow-up was doubly censored, we also estimated ψt0 relatively to the observed midpoint of infection
. We estimated its prior distribution, using simulations where we assumed a uniform probability of inclusion and dropout throughout the infection, and based on known infection duration distributions [6]. This resulted in a prior distribution of
.
These modelling choices resulted in assuming an absence of interaction between viral load time series within coinfected hosts.
Finally, for follow-ups where only one negative sample was observed at the end or at the beginning of the infection instead of two, we accounted for the risk of false-negative measurement in 10% of the cases. More specifically, we assumed ψt0 to be a mixture of the case with a true negative in 90% of the cases and a false negative in 10% of the cases, the latter leading to a different follow-up censoring status and an adapted computation of ψt0.
Parameters decomposition.
To estimate the other model parameters, we made several assumptions regarding random effects and transformation of variables. In particular, we performed a log transform for the duration parameters, and an exponential transform for the viral load to constrain it to be above v0.
Mathematically, for a given participant i, genotype g, and infection j, we estimated the following parameters:
(2A)
(2B)
where μ indicates a fixed effect, and are the host random effects,
are the genotype random effects. Ωη and Ωρ denote the variance-covariance matrices. Using the Cholesky decomposition, these were decomposed as the product of a lower triangular matrix, respectively Lη and Lρ with its conjugate transpose, and scaled elementwise by the outer product of a scale vector, with respectively
representing the variance.
In equation system 2, represents a dummy variable with value 1 when the infection is the second one with this same genotype in a given participant. Note that j, which can take values 0 or 1, is nested in g because it describes whether the second genotype is identical to the first one or not. The reverse is not true since genotype g is assumed to have the same effect across hosts. ξp and ξvl denote the reinfection (fixed) effects on the plateau duration and plateau viral load, respectively.
We did not include random effect parameters for the growth and the clearance phases of the dynamics because they were often unobserved, either due to the censoring of the follow-up or due to the short duration of those phases compared to the sampling frequency.
All the priors used and their units are listed in Table L in S1 Supplementary Materials.
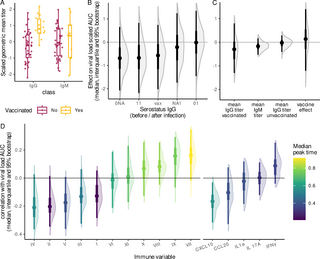
Immune dynamics: Cytokines and FCM.
Similarly, but separately, for the cytokines concentrations and leukocyte counts, we assumed an additive response due to each HPV infection, and denoted the amount of the immune marker (whether a concentration or a count) by v(t), with t the time since the beginning of the participant’s follow-up. In what follows, we will first detail the computation of v(t), and then explain its fit to the observations.
Computation of v(t).
We first decompose v(t) as a baseline value ψ0, and a specific response to each infection, z(t). Mathematically, for one infection, the value of the response against genotype g, at time t of an immune marker k can be written as follows:
(3)
where ψtm is the relative increase or decrease due to this infection, and ψdelay is the relative timing of the peak (as a proportion of the total infection duration). We used the posterior distribution of the virus dynamics to define the beginning and end of each infection, noted respectively tstart and tclear, which also correspond to the beginning and the end of the slopes (Fig 5A).
For infections with more than one genotype, we assumed an additive effect of each genotype on the immune response, assuming that the effect of each infection is independent of the others. We also assumed a baseline level for each immune marker k in each participant i, ψ0,i,k. Therefore, by summing over all the genotypes present G, we obtain the following equation for the temporal dynamics of the amount of a marker in an individual:
(4)
We corrected for this addition of several kinetics by a factor , where ncoinf is the total number of infections in a participant during the follow-up. γ was allowed to vary for each cytokine, but for identifiability reasons, we used one γFCM for all cell clusters. Intuitively, if γ = 0 each infection effect adds up, if γ = 1 immune response is the average of each infection, and if γ>1 coinfections are expected to result in a milder response than a single infection. In the case of a single infection (ncoinf = 1), the amount of a marker vi,k(t) is simply the sum of its baseline value ψ0,i,k and the deviation triggered by the infection zi,k,g(t).
From v(t) to the likelihood.
We then took into account the specificities of our samples to convert v(t) into the expected dynamics of our observations.
For the cytokines, v(t) represents the log10 of their normalised concentrations.
Due to the specificity of the tissue analysed, the flow cytometry data were compositional, meaning that their space is a simplex , with K the number of leukocytes populations. This imposed two constraints on our model.
First, we used a multinomial-logistic link between the leukocyte frequencies v′(t) and the two-slope dynamic v(t). This means that v(t) can be seen as a centered log-ratio (clr) scale of the frequencies. The clr function converts the frequencies , with K the number of leukocytes clusters, into an unbounded vector v∈ℝK with the equation:
, where g(v′) is the geometric mean of v′.
Second, since we used a clr transformation, we had to remove one degree of freedom in our equation to keep parameters identifiable. This was done by restricting the sum of the baseline level:
(5)
Similarly, to keep peak sizes identifiable, we assumed that an observed decrease in frequencies for one cell type can only be due to the increase in frequencies of other cell types (Fig 6B). Therefore, we assumed that ψtm,k>0 for all k∈FCM.
Parameters decomposition.
As for the virus kinetics, we introduced random effects and performed transformations of some variables to perform the parameter inference. In particular, we used a logit transform to constrain the peak/drop time to a proportion of the duration of the corresponding HPV infection, and a log transform for the FCM peak values to constrain them to be positive.
Mathematically, for each individual i, each HPV infecting genotype g, and each cytokine (or cell cluster) k, we estimated the following parameters:
(6A)
(6B)
(6C)
(6D)
where the μs indicate fixed effects. τ represents the correction factor for the infection duration tinf scaled,i,g so we expect the peaks early in the infection to appear proportionally later in shorter infections, and vice versa.
Furthermore, following the sum-constraint of compositional data (Eq 5), for each sub-parameter, we also have: and
.
In equation system 6, we assumed three random effects associated with each individual i. The first one, , represents the impact on the baseline value, with
. This vector is the concatenation of the effect of each cytokine and each leukocyte population:
. Joining them allows us to include a correlation between the leukocytes frequencies and the cytokine concentrations, which is biologically expected. Indeed, immune variations between women can be expected to be driven by physiological differences in body mass index (BMI), stress, or nutrition; all of which are known to affect immunity. Therefore, these could be jointly affecting the baseline level of the local immune markers that we measured. The vector is of length 15 because we have five cytokines, and ten degrees of freedom for the 11 leukocyte populations due to the sum constraint.
The other two are denoted (the random effect associated with the peak/drop value) and
(the random effect associated with the peak times shift). They are jointly correlated through a vector formed by the concatenation of (
. To avoid inflating the total number of parameters to estimate, we use the same value
for all cytokines and cell clusters of participant i, which is therefore of length 1. The underlying assumption is that the peak order is the same for all participants, following the fixed effect value, and can only be shifted earlier or later in the infection for each participant i.
There is one random effect associated to the HPV genotype g, , which is associated with the peak/drop value,
.
As for the viral kinetics model, and
denote the variance-covariance matrices and were decomposed using the Cholesky decomposition as the product of a lower triangular matrix, respectively
and
, with their conjugate transpose, and scaled elementwise by the outer product of a scale vector, respectively
and
representing variance.
Likelihood computation.
Since the five cytokine measurements originated from the same sample, we assumed that the log10 of each of their normalised concentrations, denoted ci(t), follow a multivariate normal distribution with a variance-covariance matrix Σcyt:
(7)
This matrix was also decomposed using the Cholesky decomposition, as the product of a lower triangular matrix, Lcyt with its conjugate transpose, and scaled elementwise by the outer product of a scale vector .
For the FCM, we first converted v(t) into frequencies v′(t) using the inverse of the multinomial logistic transformation, also called softmax function, i.e., .
We then used the initial cell count data, denoted fk(t) to compute the likelihood of our model. Following the results of ref. [66], we assumed a Beta-Binomial distribution to capture the counts from our compositional analyses, with a log-error parameter πk. In summary, the idea is that although the Beta-Binomial, by construction, is not constraining frequencies to sum to 1, it offers more flexibility for the error parameter which is a vector specific to each cell population k. The sum-constraint is imposed from the prediction model itself with the softmax function, which constrains the prediction vector i.e., in a simplex summing to 1.
Mathematically, this can be written as follows:
(8)
Where F is the total number of cells measured in a given sample.
The prior distributions for each parameter are described in S1 Supplementary Materials in Table M for FCM, Table N for cytokines, and Table O for the joint variance-covariance matrix.
ADVERTISEMENT:
Hai, para pengemar slot! Pernah denger semboyan “raja slot? Kalau tidak, siap-siap jatuh cinta dengan konsep ini. slot demo merupakan mesin slots yang sering kasih kemenangan. Yup, slot-slot ini bisa dikatakan sebagai jagoannya buat membawa pulang cuan. but, gimana sih
tekniknya nemuin slot demo yang benar? Santuy Bro and Sis, kita beri santai aja di tempat ini
Game tergaco saat ini hanya satu berada Indonesia hanya di akan memberikan ROI tertinggi
Daftarkanlah dengan di :
Informasi mengenai KING SLOT, Segera Daftar Bersama king selot terbaik dan terpercaya no satu di Indonesia. Boleh mendaftar melalui sini king slot serta memberikan hasil kembali yang paling tinggi saat sekarang ini hanyalah KING SLOT atau Raja slot paling gacor, gilak dan gaco saat sekarang di Indonesia melalui program return tinggi di kingselot serta pg king slot
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama kdwapp.com
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama jebswagstore.com
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama demoslotgacor.pro
slot demo gacor
slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun demo slot gacor
akun demo slot gacor permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun slot demo gacor
akun slot demo gacor permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun demo slot pragmatic
akun demo slot pragmatic permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun slot demo pragmatic
akun slot demo pragmatic permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun slot demo
akun slot demo permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
akun demo slot
akun demo slot permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
situs slot terbaru
situs slot terbaru permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
slot terbaru
slot terbaru permainan paling top dan garansi imbal balik hasil besar bersama situsslotterbaru.net
suara88 permainan paling top dan garansi imbal balik hasil besar bersama suara88.biz
sumo7777 permainan paling top dan garansi imbal balik hasil besar bersama sumo7777.com
supermoney888 permainan paling top dan garansi imbal balik hasil besar bersama supermoney888.biz
teratai88 permainan paling top dan garansi imbal balik hasil besar bersama teratai88.biz
thor88 permainan paling top dan garansi imbal balik hasil besar bersama thor88.biz
togelhk88 permainan paling top dan garansi imbal balik hasil besar bersama togelhk88.net
topjitu88 permainan paling top dan garansi imbal balik hasil besar bersama topjitu88.net
totosloto88 permainan paling top dan garansi imbal balik hasil besar bersama totosloto88.com
trisula888 permainan paling top dan garansi imbal balik hasil besar bersama trisula888.biz
udangbet88 permainan paling top dan garansi imbal balik hasil besar bersama udangbet88.net
via88 permainan paling top dan garansi imbal balik hasil besar bersama via88.biz
virusjp88 permainan paling top dan garansi imbal balik hasil besar bersama virusjp88.net
warga888 permainan paling top dan garansi imbal balik hasil besar bersama warga888.biz
waw88 permainan paling top dan garansi imbal balik hasil besar bersama waw88.biz
winjitu88 permainan paling top dan garansi imbal balik hasil besar bersama winjitu88.net
wisdom88 permainan paling top dan garansi imbal balik hasil besar bersama wisdom88.biz
wnitogel88 permainan paling top dan garansi imbal balik hasil besar bersama wnitogel88.com
yoyo888 permainan paling top dan garansi imbal balik hasil besar bersama yoyo888.biz
validtoto88 permainan paling top dan garansi imbal balik hasil besar bersama validtoto88.com
sule999 permainan paling top dan garansi imbal balik hasil besar bersama sule999.com
sule88 permainan paling top dan garansi imbal balik hasil besar bersama sule88.org
ss888bet permainan paling top dan garansi imbal balik hasil besar bersama ss888bet.com
sia77 permainan paling top dan garansi imbal balik hasil besar bersama sia77.info
seluang88 permainan paling top dan garansi imbal balik hasil besar bersama seluang88.com
satu88 permainan paling top dan garansi imbal balik hasil besar bersama satu88.biz
satu777 permainan paling top dan garansi imbal balik hasil besar bersama satu777.asia
rp88 permainan paling top dan garansi imbal balik hasil besar bersama rp88.biz
rp88 permainan paling top dan garansi imbal balik hasil besar bersama rp88.asia
rp88 permainan paling top dan garansi imbal balik hasil besar bersama rp77.live
qiuqiu88 permainan paling top dan garansi imbal balik hasil besar bersama qiuqiu88.biz
pt88 permainan paling top dan garansi imbal balik hasil besar bersama pt88.org
pt77 permainan paling top dan garansi imbal balik hasil besar bersama pt77.info
produk88 permainan paling top dan garansi imbal balik hasil besar bersama produk88.asia
mt88 permainan paling top dan garansi imbal balik hasil besar bersama mt88.org
mt77 permainan paling top dan garansi imbal balik hasil besar bersama mt77.biz
menang66 permainan paling top dan garansi imbal balik hasil besar bersama menang66.biz
latobet888 permainan paling top dan garansi imbal balik hasil besar bersama latobet888.org
kedai96 permainan paling top dan garansi imbal balik hasil besar bersama kedai96.org
kedai188 permainan paling top dan garansi imbal balik hasil besar bersama kedai188.biz
ids88 permainan paling top dan garansi imbal balik hasil besar bersama ids88.biz
hp88 permainan paling top dan garansi imbal balik hasil besar bersama hp88.org
hp77 permainan paling top dan garansi imbal balik hasil besar bersama hp77.org
gm88 permainan paling top dan garansi imbal balik hasil besar bersama gm88.asia
gm77 permainan paling top dan garansi imbal balik hasil besar bersama gm77.net
final888 permainan paling top dan garansi imbal balik hasil besar bersama final888.org
duit88 permainan paling top dan garansi imbal balik hasil besar bersama duit88.asia
duit168 permainan paling top dan garansi imbal balik hasil besar bersama duit168.biz
divisi88 permainan paling top dan garansi imbal balik hasil besar bersama divisi88.org
dewi500 permainan paling top dan garansi imbal balik hasil besar bersama dewi500.biz
devil88 permainan paling top dan garansi imbal balik hasil besar bersama devil88.info
cuputoto88 permainan paling top dan garansi imbal balik hasil besar bersama cuputoto88.com
cukongbet88 permainan paling top dan garansi imbal balik hasil besar bersama cukongbet88.asia
bom888 permainan paling top dan garansi imbal balik hasil besar bersama bom888.biz
bintaro888 permainan paling top dan garansi imbal balik hasil besar bersama bintaro888.info
askasino88 permainan paling top dan garansi imbal balik hasil besar bersama askasino88.org
999aset permainan paling top dan garansi imbal balik hasil besar bersama 999aset.com
afb77 permainan paling top dan garansi imbal balik hasil besar bersama afb77.biz
aset99 permainan paling top dan garansi imbal balik hasil besar bersama aset99.biz
bendera77 permainan paling top dan garansi imbal balik hasil besar bersama bendera77.biz
bendera888 permainan paling top dan garansi imbal balik hasil besar bersama bendera888.com
coco88 permainan paling top dan garansi imbal balik hasil besar bersama coco88.org
cuma77 permainan paling top dan garansi imbal balik hasil besar bersama cuma77.biz
cuma88 permainan paling top dan garansi imbal balik hasil besar bersama cuma88.org
dwv88 permainan paling top dan garansi imbal balik hasil besar bersama dwv88.org
fafajp88 permainan paling top dan garansi imbal balik hasil besar bersama fafajp88.com
gemar88 permainan paling top dan garansi imbal balik hasil besar bersama gemar88.biz
gocap88 permainan paling top dan garansi imbal balik hasil besar bersama gocap88.info
gocaptoto permainan paling top dan garansi imbal balik hasil besar bersama gocaptoto.asia
hakabet88 permainan paling top dan garansi imbal balik hasil besar bersama hakabet88.com
hwtoto88 permainan paling top dan garansi imbal balik hasil besar bersama hwtoto88.org
ina77 permainan paling top dan garansi imbal balik hasil besar bersama ina77.biz
ina88 permainan paling top dan garansi imbal balik hasil besar bersama ina88.info
jingga8888 permainan paling top dan garansi imbal balik hasil besar bersama jingga8888.com
juragan777 permainan paling top dan garansi imbal balik hasil besar bersama juragan777.asia
kastil77 permainan paling top dan garansi imbal balik hasil besar bersama kastil77.info
kebo888 permainan paling top dan garansi imbal balik hasil besar bersama kebo888.biz
kkwin77 permainan paling top dan garansi imbal balik hasil besar bersama kkwin77.com
kokoslot88 permainan paling top dan garansi imbal balik hasil besar bersama kokoslot88.asia
luckydf88 permainan paling top dan garansi imbal balik hasil besar bersama luckydf88.org
microstar888 permainan paling top dan garansi imbal balik hasil besar bersama microstar888.biz
monperatoto88 permainan paling top dan garansi imbal balik hasil besar bersama monperatoto88.com
mpo1122 permainan paling top dan garansi imbal balik hasil besar bersama mpo1122.biz
mpo122 permainan paling top dan garansi imbal balik hasil besar bersama mpo122.biz
mpopelangi88 permainan paling top dan garansi imbal balik hasil besar bersama mpopelangi88.com
pamanslot88 permainan paling top dan garansi imbal balik hasil besar bersama pamanslot88.biz
panel88 permainan paling top dan garansi imbal balik hasil besar bersama panel88.org
paragon77 permainan paling top dan garansi imbal balik hasil besar bersama paragon77.biz
paragon888 permainan paling top dan garansi imbal balik hasil besar bersama paragon888.info
pion77 permainan paling top dan garansi imbal balik hasil besar bersama pion77.biz
prada88 permainan paling top dan garansi imbal balik hasil besar bersama prada88.asia
prada888 permainan paling top dan garansi imbal balik hasil besar bersama prada888.com
qqslot88slot permainan paling top dan garansi imbal balik hasil besar bersama qqslot88slot.com
rejekibet88 permainan paling top dan garansi imbal balik hasil besar bersama rejekibet88.com
rezekibet88 permainan paling top dan garansi imbal balik hasil besar bersama rezekibet88.org
sensa77 permainan paling top dan garansi imbal balik hasil besar bersama sensa77.biz
sensa888 permainan paling top dan garansi imbal balik hasil besar bersama sensa888.biz
singajp88 permainan paling top dan garansi imbal balik hasil besar bersama singajp88.com
sr77 permainan paling top dan garansi imbal balik hasil besar bersama sr77.org
sr88 permainan paling top dan garansi imbal balik hasil besar bersama sr88.org
surya77 permainan paling top dan garansi imbal balik hasil besar bersama surya77.biz
surya88 permainan paling top dan garansi imbal balik hasil besar bersama surya88.asia
tajir77 permainan paling top dan garansi imbal balik hasil besar bersama tajir77.info
tajir88 permainan paling top dan garansi imbal balik hasil besar bersama tajir88.biz
toto122 permainan paling top dan garansi imbal balik hasil besar bersama toto122.com
toto123 permainan paling top dan garansi imbal balik hasil besar bersama toto123.biz
uangvip88 permainan paling top dan garansi imbal balik hasil besar bersama uangvip88.com
wajik77 permainan paling top dan garansi imbal balik hasil besar bersama wajik77.asia
777neko permainan paling top dan garansi imbal balik hasil besar bersama 777neko.org
88judi permainan paling top dan garansi imbal balik hasil besar bersama 88judi.net
99judi permainan paling top dan garansi imbal balik hasil besar bersama 99judi.org
abcslot88 permainan paling top dan garansi imbal balik hasil besar bersama abcslot88.asia